Writing Efficient R Code
One basic thing to do is to keep the R version up to date, since every update aims to make some processes more efficient. The version command returns a list that contains (among other things) the major and minor version of R currently being used. 
The Art of Benchmarking
Is my code really slow? We need to compare the current solution with one or more alternatives. We just calculate the time each one takes to do the same, and we select the fastest.
Benchmarking:
- We construct a function around the feature we wish to benchmark
- We time the function under different scenarios, e.g data set
Timing with system.time
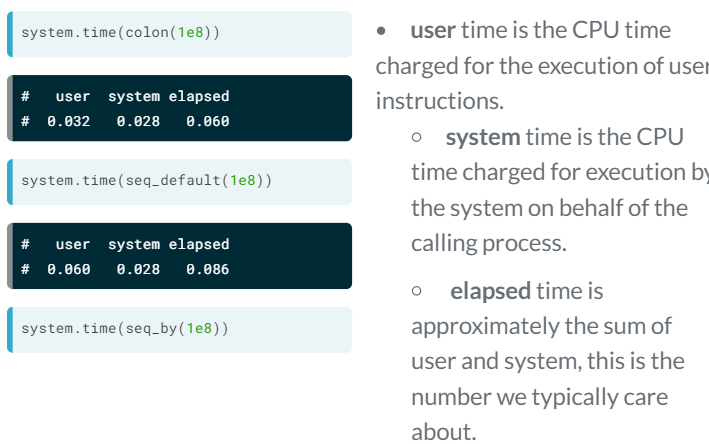
Storing the result
Microbenchmark package

- Comparing read times of CSV and RDS. One of the most common tasks we perform is reading in data from CSV files. However, for large CSV files this can be slow. One neat trick is to read in the data and save as an R binary file (
rds) usingsaveRDS(). To read in therdsfile, we usereadRDS().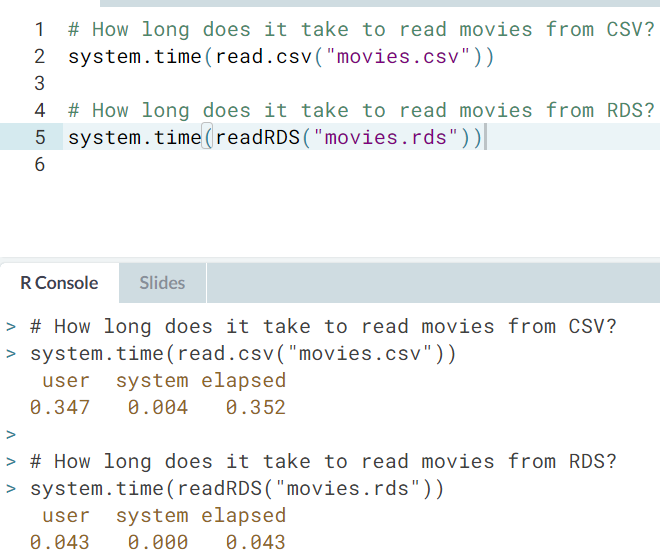
How good is your machine
Of course the speed of an operation to be done depends on the power of the computer. You can compare the speed of your computer in doing some standard tasks with the Bechmarkme package, both in absoute time of such tasks and relatively to each other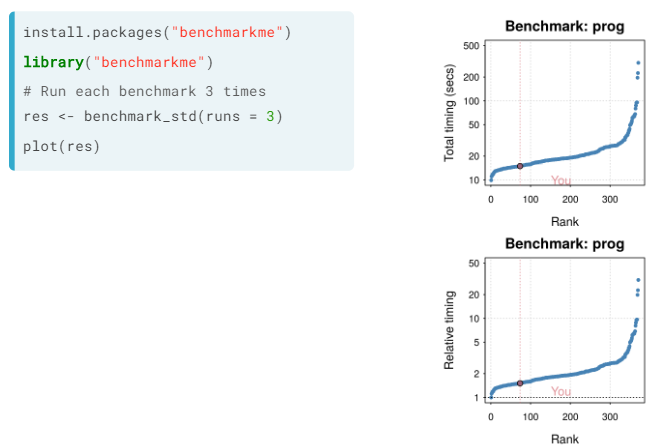
Fine Tuning: Efficient Base R
R is flexible because you can often solve a single problem in many different ways. Some ways can be several orders of magnitude faster than the others. This chapter teaches you how to write fast base R code.
Memory allocation
- In R, memory allocation happens automatically.
- R allocates memory in RAM to store variables.
- Minimize variable assignment for speed.
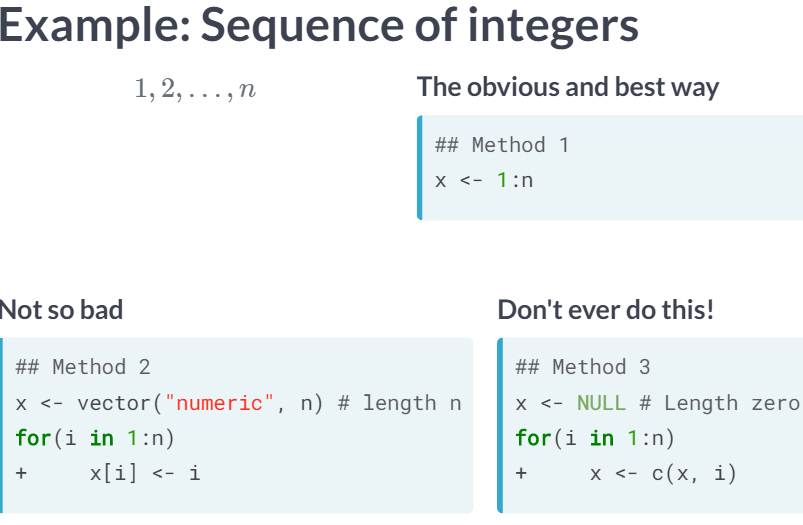
Ex. 1 allocates the vector directly, 2 preallocates the vector, and 3 grows the vector
 |  |
Importance of vectorizing your code
When we call a base R function, we eventually call a C or FORTRAN code, and that underlying code is very heavily optimised. So to make R code faster is to a access this optimised code as quickly as possible: This usually means vectorised code.
Vectorised functions. Many R functions are vectorised. Ex: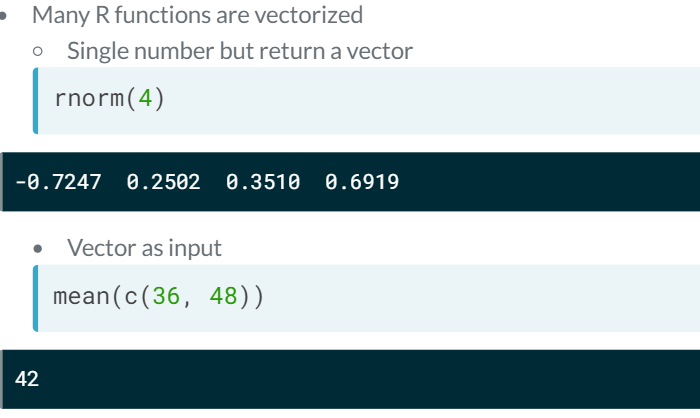
- Why is the loop slow?
Let's supose we want to obtain a million standard normal distributed numbers with these two aproaches. In the loop we are calling the function everytime we enter while in the allocated one only one.
Data frames and matrices
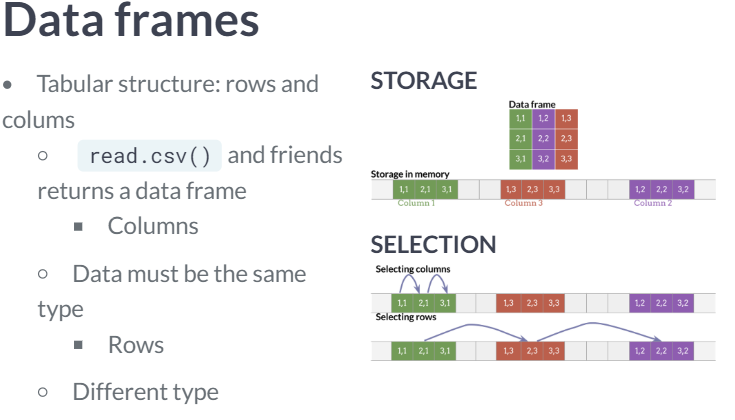 | 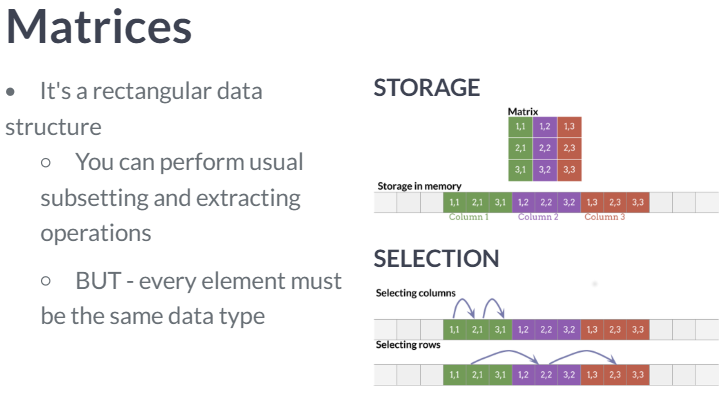 |
The substantial difference is that data frames are multiple vectors while matrices are a single vector.
- Because all values in a matrix object must be the same data type, it is much faster to access the first column of a matrix than it is to access that of a data frame.
- The most notable time consuming task of data frames wrt matrices is the row selection.
Code Profiling
Profiling helps you locate the bottlenecks in your code (which part is the one that slows down the overall process). This chapter teaches you how to visualize the bottlenecks using the `profvis` package. 
profvis package
There are two ways of profiling with this package.
- Integrated support from RStudio
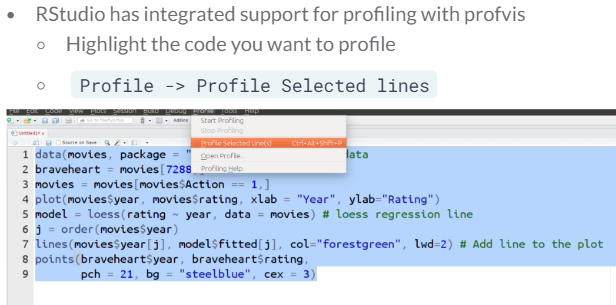
- Using the command line

And the resulting output for the function call is the following, which gives the space and time required for each function of the code.
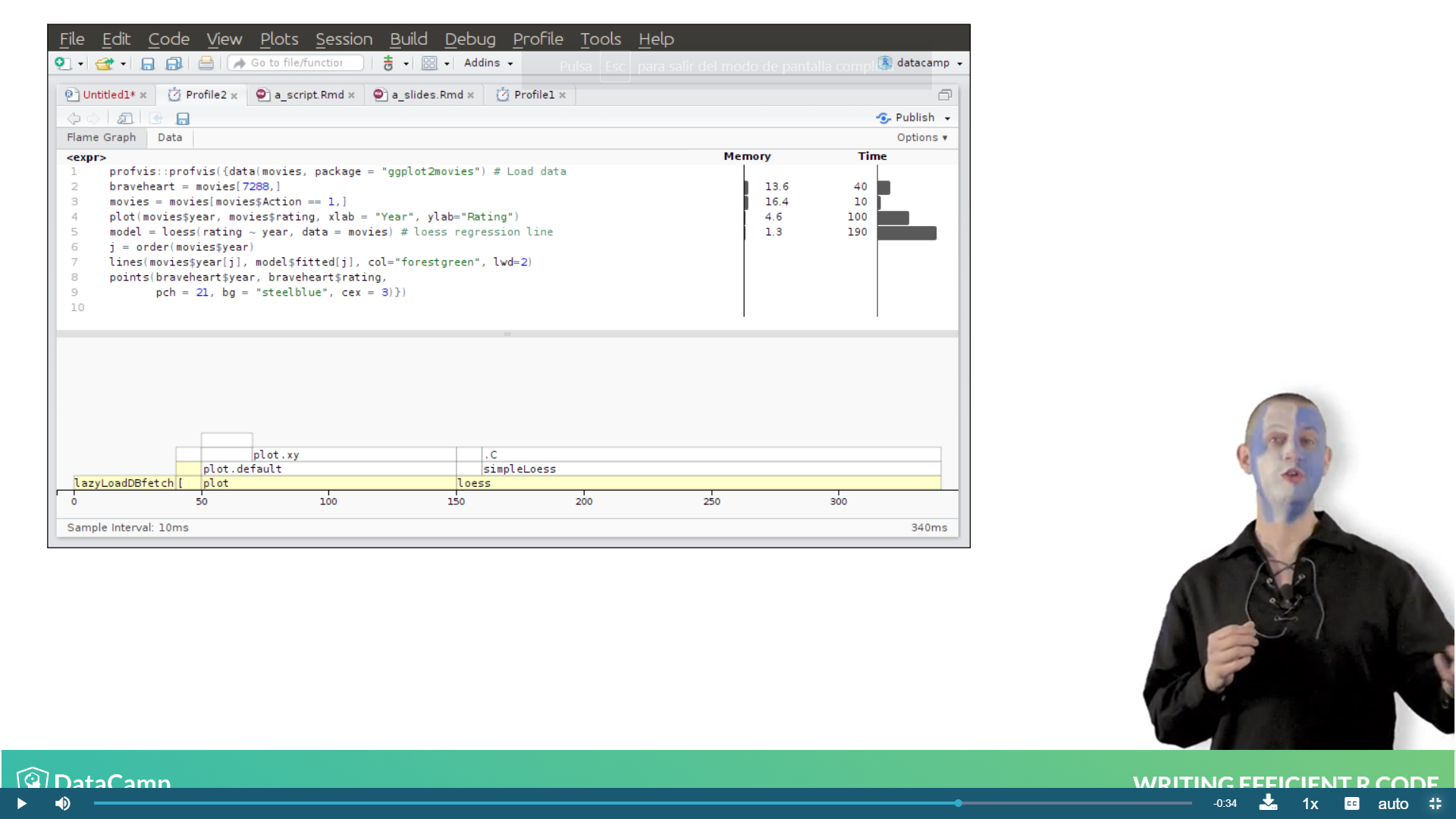
Ex. The monopoly simulation.
- Change the data frame to a matrix. Where we generated the possible dice rolls and stored the results in a data frame:
df <- data.frame(d1 = sample(1:6, 3, replace = TRUE), d2 = sample(1:6, 3, replace = TRUE))
We can optimize this code by making two improvements:- Switching from a data frame to a matrix
- Generating the 6 dice rolls in a single step
This givesm <- matrix(sample(1:6, 6, replace = TRUE), ncol = 2)
- Calculating row sums. The second bottleneck identified was calculating the row sums.
total <- apply(d, 1, sum)In the previous exercise you switched the underlying object to a matrix. This makes the above apply operation three times faster. But there's one further optimization you can use - switchapply()withrowSums(). - Use && instead of &. The
&operator will always evaluate both its arguments. That is, if you typex & y, R will always try to work out whatxandyare. The&&operator takes advantage of this trick, and doesn't bother to calculateyif it doesn't make a difference to the overall result. One thing to note is that&&only works on single logical values but&also works on vectors of length greater than 1.
Parallel Programming
Some problems can be solved faster using multiple cores on your machine.
- CPUs. By default, R only uses one core, so we use the "parallel" package so that different parts of our code are ran in each core
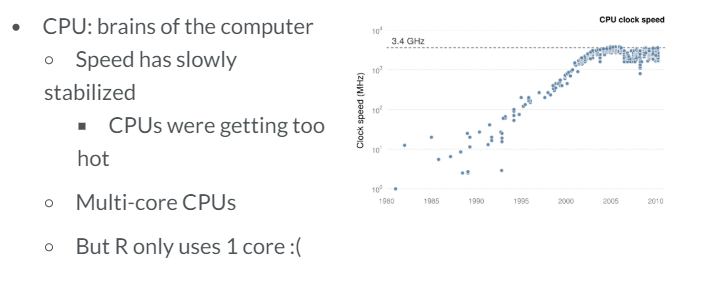
- Your CPU. These functions may be used to determine our CPU number of cores
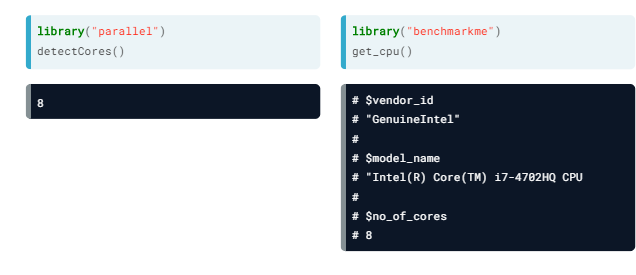
- Your CPU. These functions may be used to determine our CPU number of cores
- What can be run in parallel? Many statistical algorithms just haven't been designed with parallel programming in mind; this is starting to change, but most are not designed for that.
- Ex. Can be done in parallel

- Ex. Cannot be done in parallel

- The general idea is that parallel programming can be done on loops on which elements of the loop are mutually independent
- Ex. Can be done in parallel
Parallel packages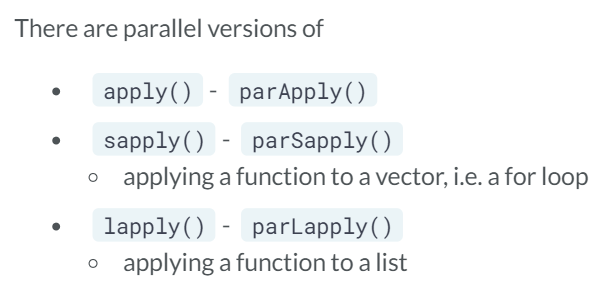
- The parallel package - parApply. If we intend to use our computer for something else apart from our R computation, we specify the number of cores to use to be the total minus one.

- How would we do this in parallel?
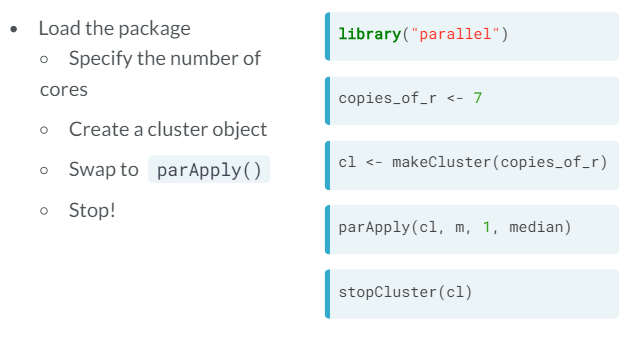
- makeCluster creates copies of R running in parallel
- To take into consideration. Sometimes running in parallel is slower due to thread communication, so it's a good idea to benchmark both solutions.
- How would we do this in parallel?
- The parallel package - parSapply.

To run in parallel we need the same recipe: Load the package, make a cluster, sitch to parSapply(), and stop the cluster.- Ex. Bootstrapping.
 We detect there is a correlation between attack and defense in our Pokemon dataset. First we create our bootstrap function
We detect there is a correlation between attack and defense in our Pokemon dataset. First we create our bootstrap function We have to run multiple times this function, and that's where parSapply enters.
We have to run multiple times this function, and that's where parSapply enters.
- Note that, unlike parApply, here we need to export functions/data before calling parSapply; it is done like this for optimization.
- As we mentioned earlier, the communication between cores has a computational cost, so depending on the problem is that parallel computation is worth it.
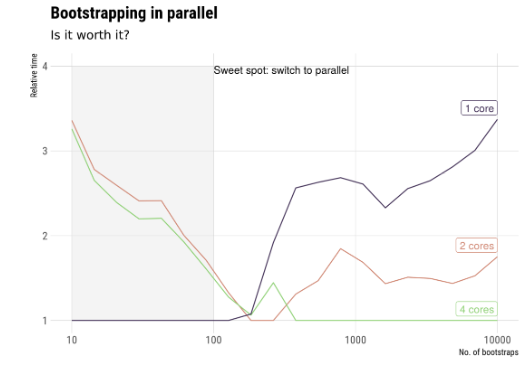
- Ex. Bootstrapping.



